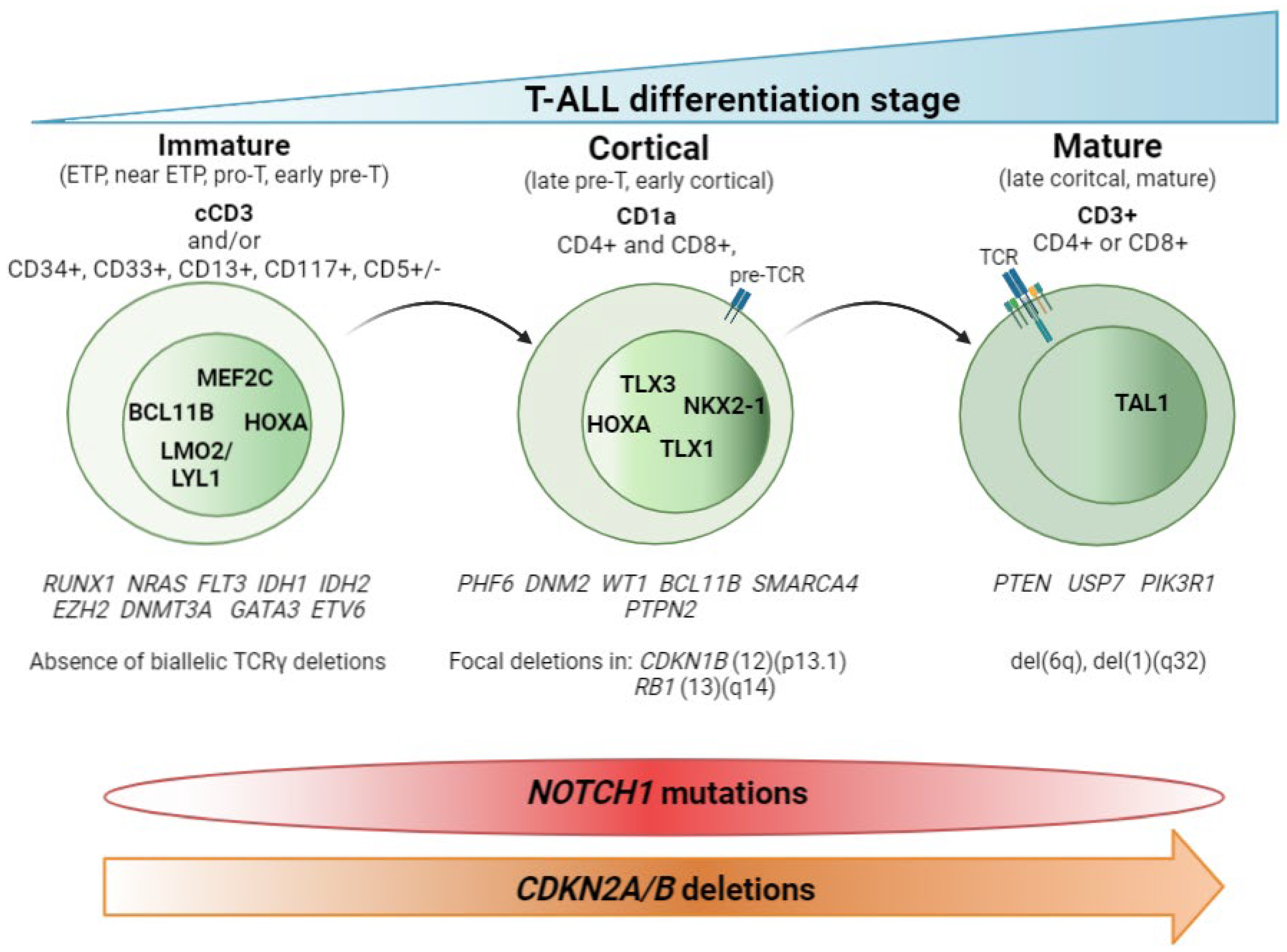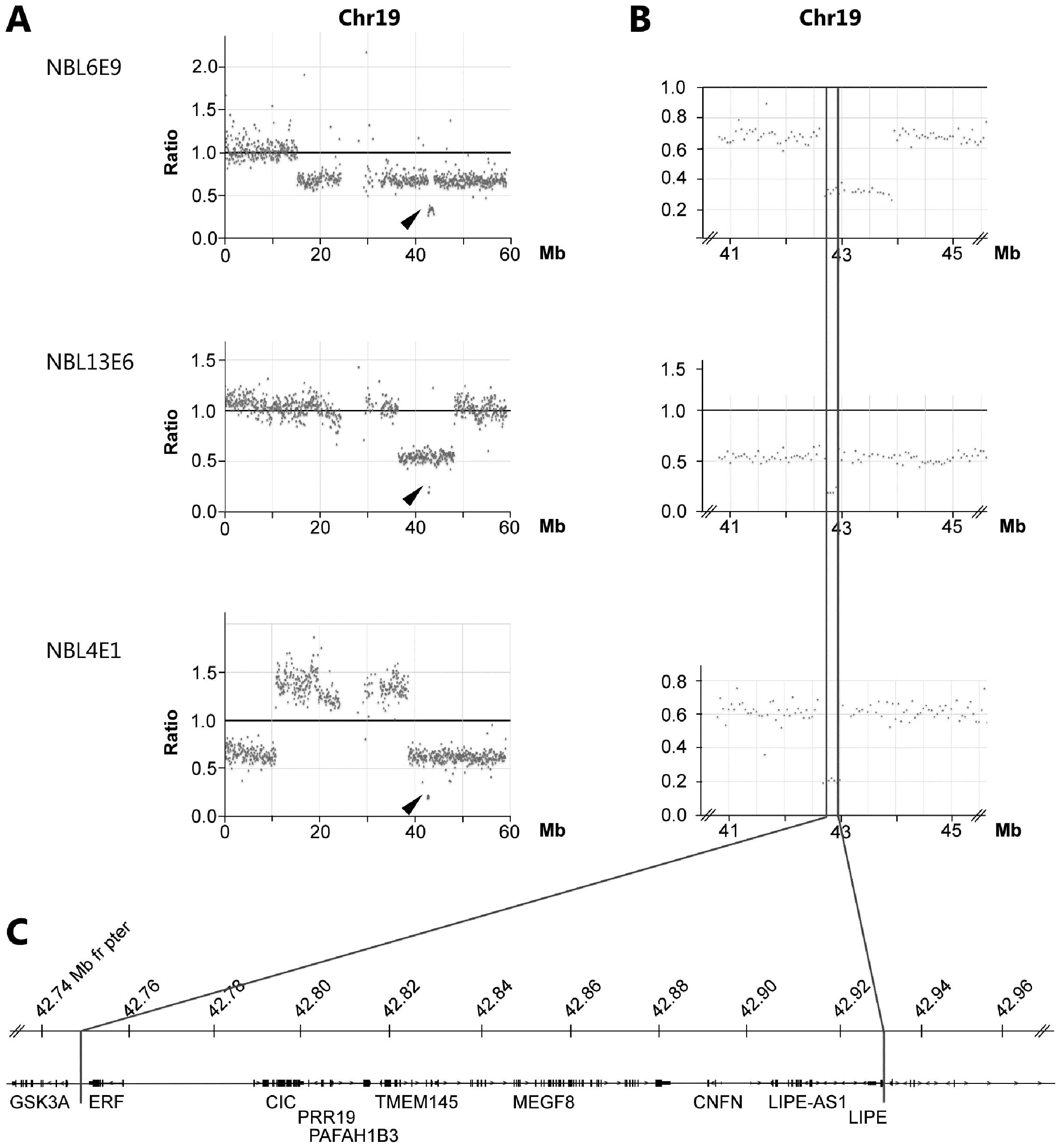
Estimation of copy number aberrations: Comparison of exome sequencing data with SNP microarrays identifies homozygous deletions of 19q13.2 and CIC in neuroblastoma
The PAX5 gene is frequently rearranged in BCR-ABL1-positive acute lymphoblastic leukemia but is not associated with outcome. A report on behalf of the GIMEMA Acute Leukemia Working Party | Haematologica
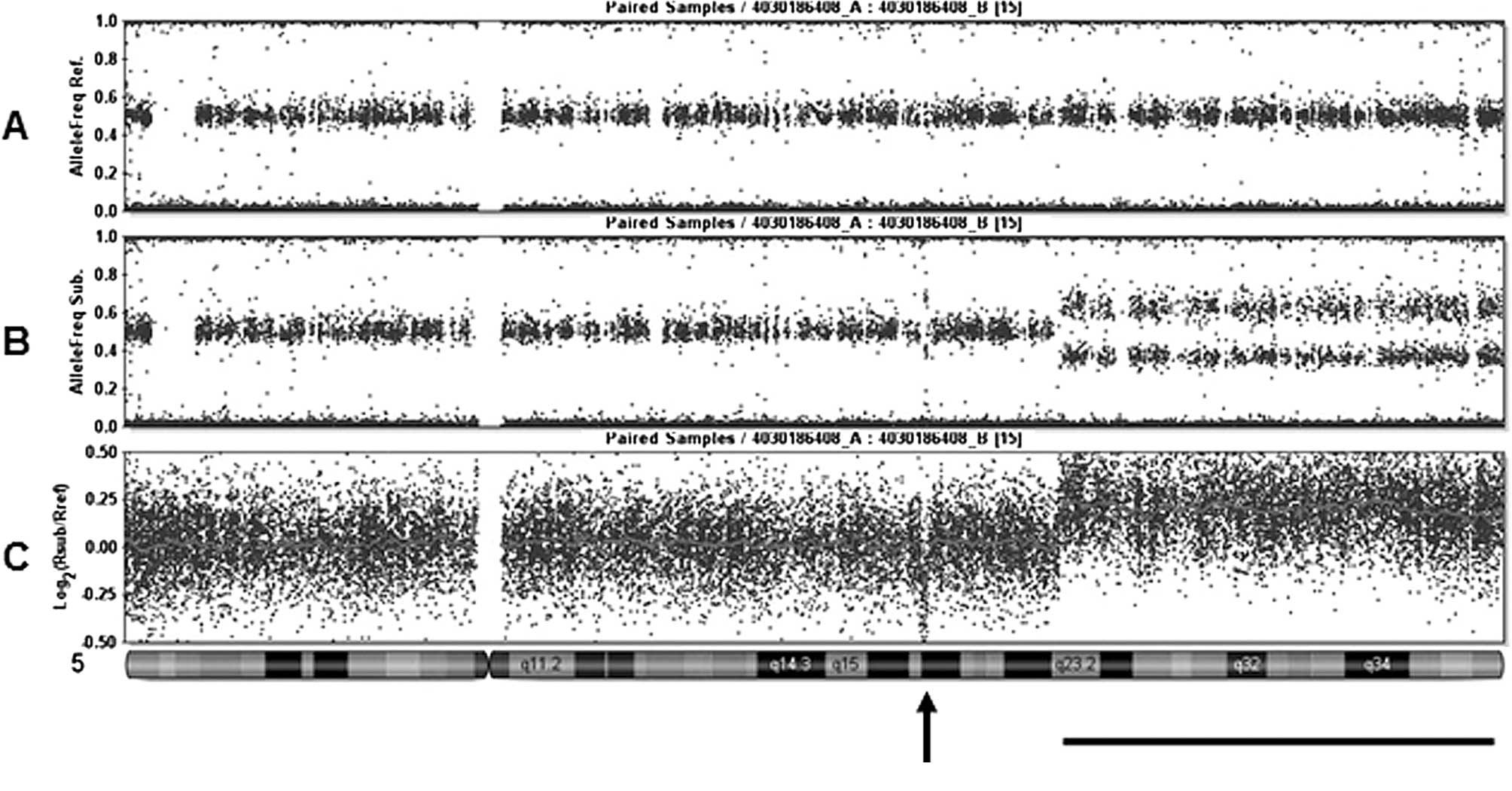
Histopathologic subtype-specific genomic profiles of renal cell carcinomas identified by high-resolution whole-genome single nucleotide polymorphism array analysis
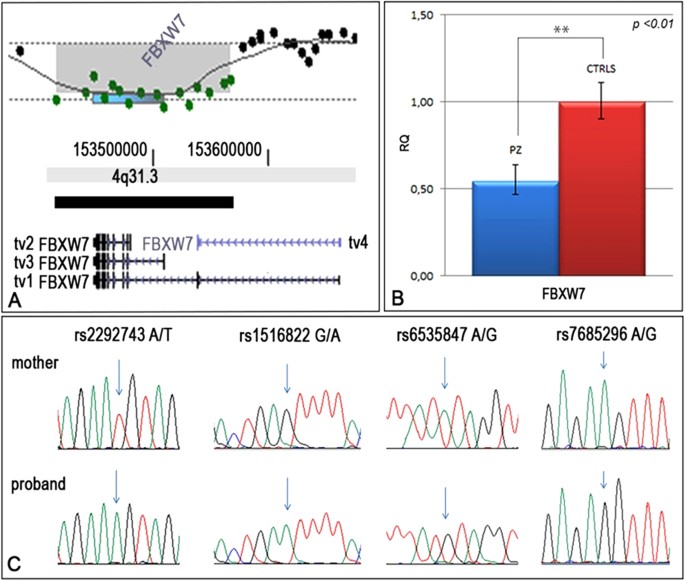
Constitutional de novo deletion of the FBXW7 gene in a patient with focal segmental glomerulosclerosis and multiple primitive tumors | Scientific Reports
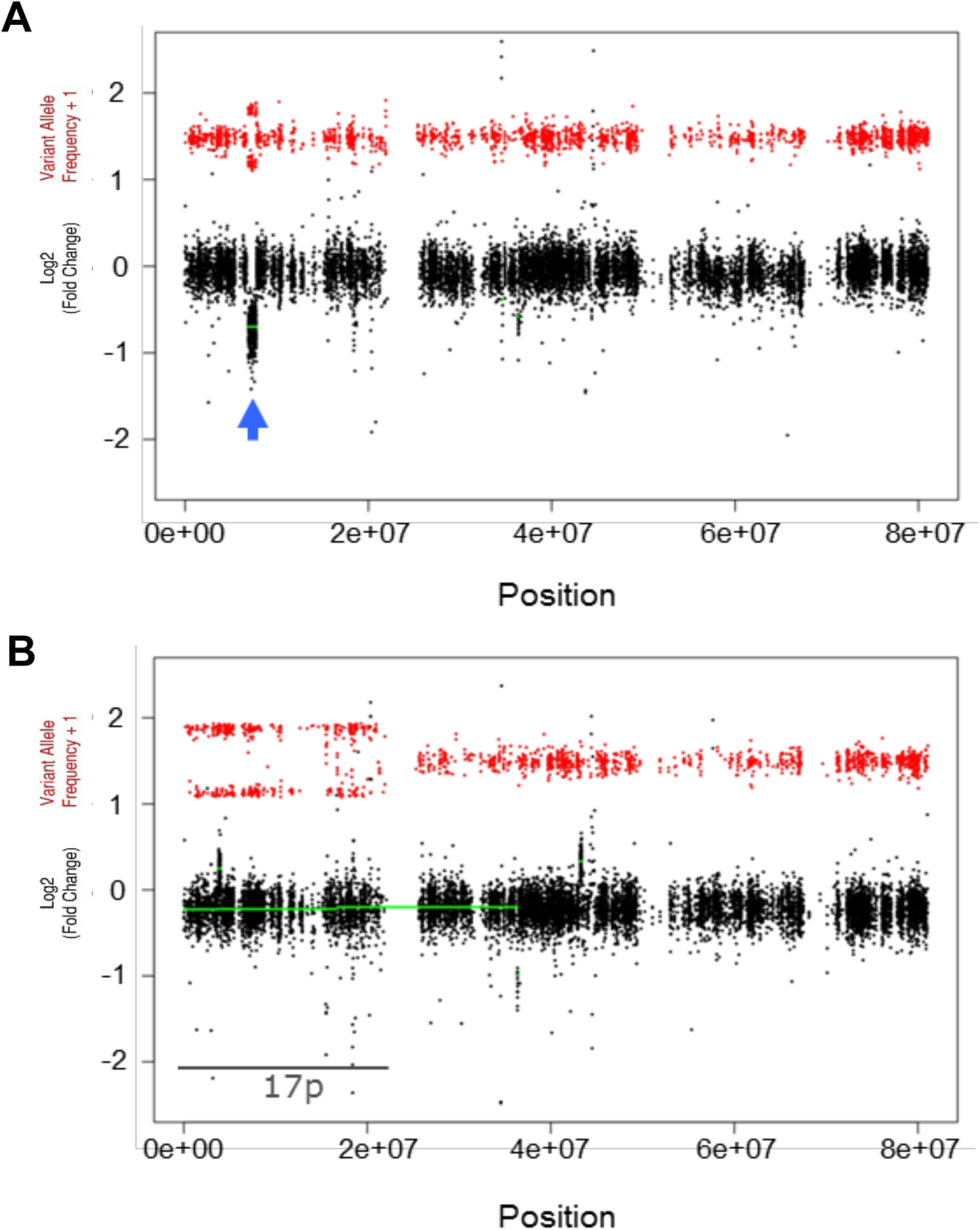
CNV Radar: an improved method for somatic copy number alteration characterization in oncology | BMC Bioinformatics | Full Text

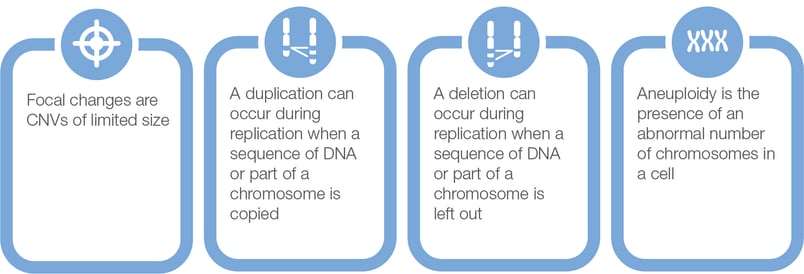
Sites in the genome of frequent cancer focal deletions. (A) Average DNA... | Download Scientific Diagram

Focal Deletion of the Adenosine A1 Receptor in Adult Mice Using an Adeno-Associated Viral Vector | Journal of Neuroscience

Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma | PNAS
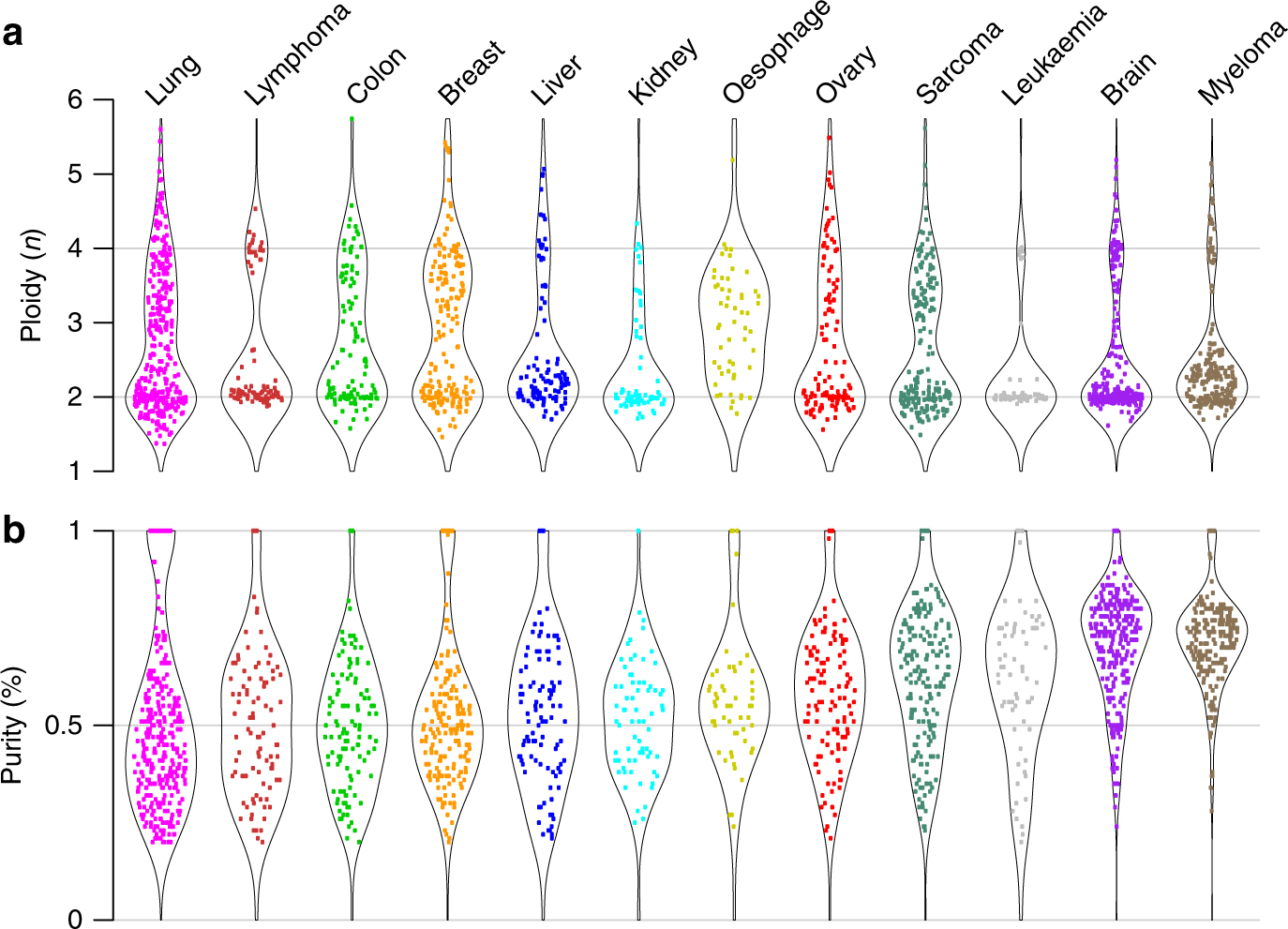
Pan-cancer analysis of homozygous deletions in primary tumours uncovers rare tumour suppressors | Nature Communications
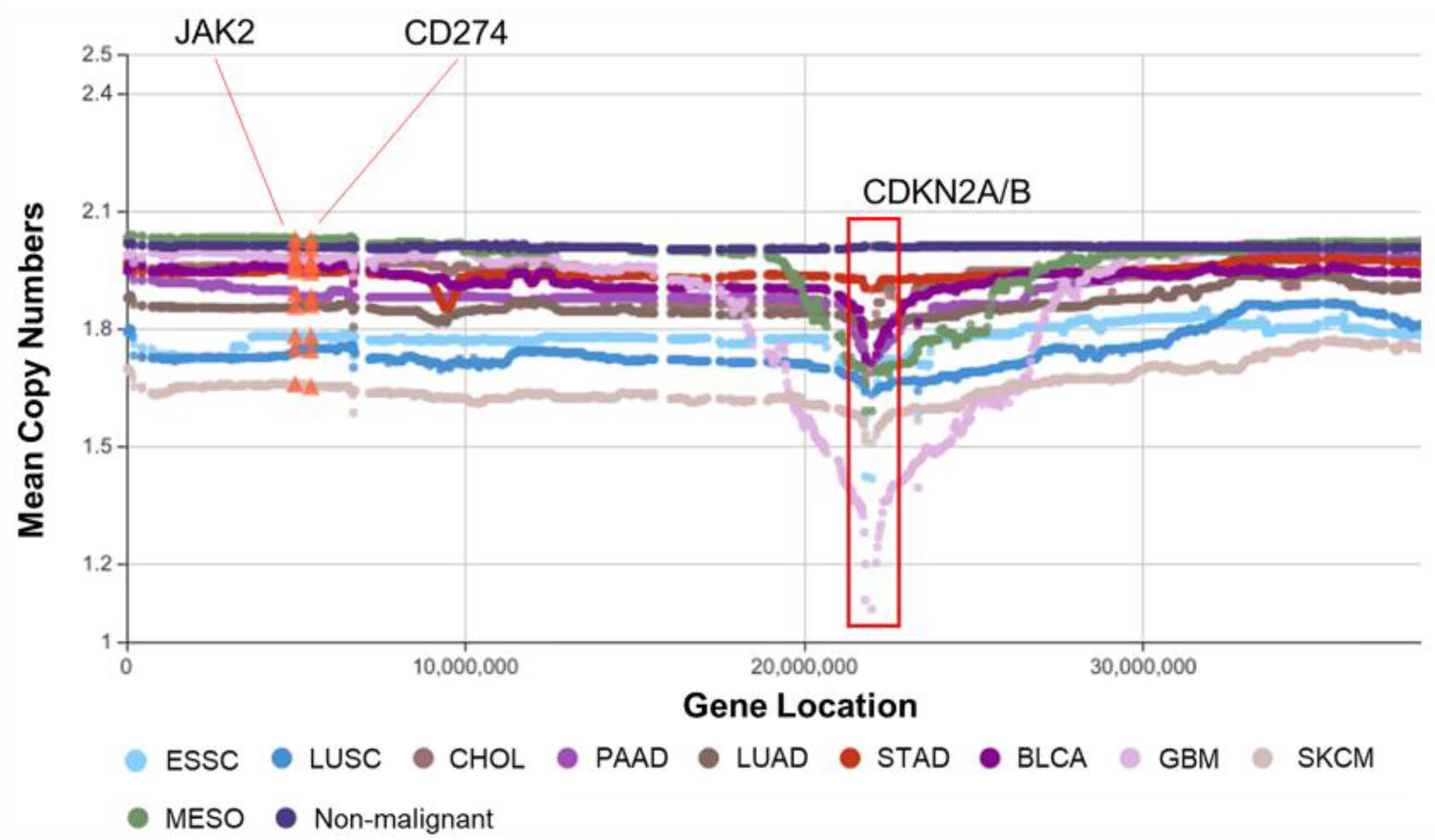
Symmetry | Free Full-Text | Machine Learning Reveals Molecular Similarity and Fingerprints in Structural Aberrations of Somatic Cancer

Genome-wide somatic copy number alteration analysis and database construction for cervical cancer | SpringerLink

The focal deletion of chr6p21.32 encompassing HLA class II alleles. a... | Download Scientific Diagram
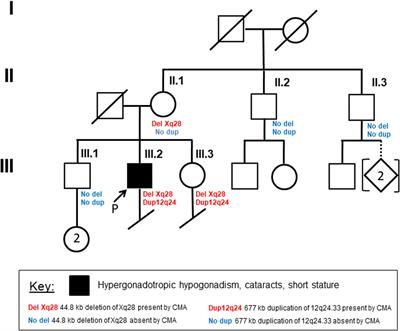
Frontiers | Deletion of FUNDC2 and CMC4 on Chromosome Xq28 Is Sufficient to Cause Hypergonadotropic Hypogonadism in Men